Normalization
Normalization in WebIDQ adjusts study sample concentrations based on the observed concentration changes in a reference sample. The normalization can be performed within a plate or across series of plates to improve accuracy and reduce batch effects in sample data.
Requirements
For data normalization of study samples, a biocrates QCs or a suitable study reference sample can be used. In order to guarantee a robust data normalization, it is recommended to analyze biocrates QC level 2 in replicates of at least 4 per plate. To perform a data normalization load each plate-run in Results. Be aware that data normalization will change the displayed concentration values. Data normalization can be undone by deactivating the toggle next to the Data Normalization ▾ dropdown menu in Results.

Normalization for kit results
Types of normalization
- Normalize sample concentration data: Check this box to activate normalization for the active sample data. A sample source must be selected as the reference sample. Only samples that are detected with a minimum of 3 replicates per plate will be available for selection.
- Normalization vs. target values: Metabolites are normalized to correct for deviation in accuracy observed in the quality control (QC) samples (only available when using biocrates QC samples for normalization). When checked, intra-plate normalization is performed (required when using the Quant 500 kit). Unchecked, normalization will only apply to inter-plate samples by adjusting for the average variation in the reference sample.
Metabolite classifications
- 7-point calibrated metabolites: normalization is not required, but recommended. Normalization may be required, if the derivatization performance is inconsistent or calibration curves show different shapes between kit runs.
- 1-point calibrated metabolites: normalization is required for all metabolites.
Suggested normalization procedures
Target normalization using QC2 as a reference sample is recommended for kits that contain 1 point calibrated metabolites such as Quant 500 (XL), p400 HR, Quant HR Xpress, and p180 kits.
QC normalization is not required for Bile Acids kit, as all metabolites utilize a 7 point calibration. However, in some circumstances, it may be used to improve inter-plate performance.
Choosing a sample for normalization
Normalization can be performed using either biocrates QC samples or a Custom QC. The choice of normalization reference sample will depend on the study goals and nature of the experimental samples. Below are rationale and suggestions for each of the choices.
biocrates offers expert support for statistical analysis. Please contact us for more information.
Using biocrates QC2 as a reference sample is straightforward and provides several benefits as a normalization option:
- biocrates QCs are plasma-based samples included with each kit.
- Intra-plate normalization can be performed via target value normalization.
- QCs from the same or different batches can be used.
- QC2 concentrations provide a reasonable compromise for all metabolites.
Depending on the metabolite concentrations present in the study samples, a more robust normalization can be performed by utilizing different QC levels for specific metabolites. For example:
- Average concentrations of metabolites A, B, C are in the range of corresponding concentrations in QC2 - use QC2 for normalization procedure of A, B, C.
- Average concentrations of metabolites X, Y, Z are in the range of corresponding concentrations in QC1 - use QC1 for normalization procedure of X, Y, Z.
For optimal normalization performance or when working in sample matrices other than plasma, select a study pool as reference sample, representing the average concentrations of the experimental samples.
- The study pool is specific to the current project. This sample may not be available for future projects, limiting the chance to perform inter-plate normalizations across results from long-term studies.
- If normalization of results across different studies is required, a larger external sample pool should be generated.
- Samples from this external reference pool should be included in the present and all future studies for normalization.
Target value normalization
Data normalization based on QC target values applies the bias between measured and expected metabolite concentration in QC samples. The target value normalization is possible only if QC samples of the same QC Level are present on all selected plate-runs. Only when biocrates QC samples are selected as the sample source can WebIDQ perform target value normalization.
Algorithm in details:
- For every plate-run, the user chooses one QC sample as the source sample. (They must all be the same QC Level).
- The mean or median concentration (specified by user) of all measured concentration values in the source samples is calculated for every combination of plate-run and metabolite.
- The best matching (according OP Restriction) expected concentration is obtained from the source samples for every combination of plate-run and metabolite.
- Every metabolite concentration measured in every sample is divided by the mean/median concentration (step 2) and multiplied by the expected concentration (step 3) of the same plate-run and metabolite.
Only measurements having a nonzero value, status other than "invalid", not "excluded" by user, and greater than LOD are considered for normalization.
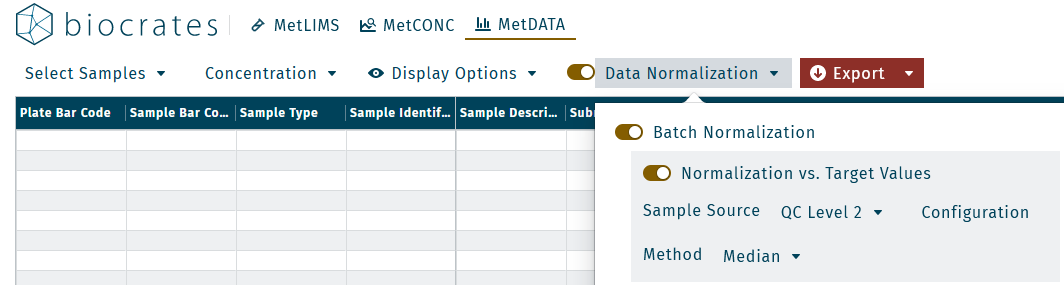
- Activate Data normalization in the Results subheader.
- Select a Plate source for normalization.
- All: average values of all linked plates are used for normalization.
- Specific plate: values of the selected plate are used as reference for normalization.
- Select a biocrates QC in the Sample source field.
- Activate the option Normalize vs. target values.
- For average concentration calculations of replicates, select the algorithm median or mean for data normalization in the Method field. We recommend using the median.
Data Normalization does not overwrite original kit plate concentration data. To perform normalization using non-biocrates QC samples, refer to the section Reference sample normalization.
The following steps describe how the data normalization is carried out by WebIDQ:
- Analyte by analyte and for every kit plate, the median or mean values (if above LOD) of the selected sample source are calculated, hereinafter referred to as A. For plate 1 the value A1 is calculated, for plate 2 the value A2, and so on.
- Per analyte and per QC level batch, one concentration Target Value (TV) is available in the WebIDQ database, e. g. TVb1 for QC batch 1.
- The ratio Ab1/TVb1 is calculated and represents the correction factor for each kit plate where QC batch 1 was used, hereinafter referred to as Cb1.
In detail, C1b1 = A1b1/TVb1 is the correction factor for an analyte on kit plate 1, C2b1 = A2b1/TVb1 is the correction factor for an analyte on kit plate 2, and so on. The metabolite concentrations are finally normalized by dividing each concentration value by the correction factor Cb1, e.g. c(analyte, plate1)normalized = c(analyte, plate1) / C1b1(analyte).
biocrates QCs from different production batches can be used with target value normalization.
Reference sample normalization
Data normalization based on reference sample eliminates the measurement bias between plates. The reference sample normalization is possible only if at least one QC or Unknown sample is present on all selected plate-runs. Only when a suitable reference sample is selected as the sample source can WebIDQ perform target value normalization.
Algorithm in detail:
- The user chooses one QC or Unknown sample as the source sample. The samples must be present on all selected plate-runs.
- The user optionally chooses one particular plate-run as the source plate. If none is chosen, all plate-runs are considered to be source plates.
- The mean or median concentration (specified by user) of all measured concentration values in the source sample is calculated for every combination of plate-run and metabolite - the plate-run specific mean/median. The mean/median of all measured concentration values** in the source samples on the source plate(s) is calculated per every metabolite - the overall mean/median.
- Every metabolite concentration measured in every sample is divided by the plate-run specific mean/median (step 3) and multiplied by the overall mean/median (step 4) of the respective plate-run and metabolite.
Only measurements having a nonzero value, status other than "invalid", not "excluded" by user, and greater than LOD are considered for normalization.
For data normalization of unknown samples, results of a QC sample can be used as reference. In order to guarantee a robust data normalization, we recommend analyzing biocrates QC level 2 or your own QC sample in replicates, at least of 4. To perform a data normalization, select and link all biocrates QC and unknown samples of the kit plates in Results > Select samples. Be aware that data normalization will change the displayed concentration values. Data normalization can be undone by deactivating the toggle next to the Data normalization ▾ dropdown menu in Results.
- Activate Data normalization in the Results submenu.
- Select a Plate source for normalization.
- All: average values of all linked plates are used for normalization.
- Specific plate: values of the selected plate are used as reference for normalization.
- Select either a biocrates QC or one of your samples in the Sample source field.
- Deactivate the option Normalize vs. target values.
- For average concentration calculations of replicates, select the algorithm median or mean for data normalization in the Method field. We recommend using the median.
Data normalization does not alter original kit plate concentration data. Any sample that was measured in all loaded plate-runs with the appropriate number of replicates can be used as sample source for data normalization. This offers the flexibility to use your own QC or pooled samples.
The following steps describe how the data normalization is carried out by WebIDQ:
- Analyte by analyte and for every kit plate, the median or mean values (if above LOD) of the selected sample source is calculated, hereinafter referred to as A. For plate 1 the value A1 is calculated, for plate 2 the value A2, and so on. If the selected sample source has been pipetted only once onto a kit plate, there is only one value per analyte.
- Analyte by analyte, the overall median or mean value across all selected plates is calculated, hereinafter referred to as B.
- The ratio A / B is calculated and represents the correction factor for each kit plate, hereinafter referred to as C. In detail, C1 = A1 / B is the correction factor for an analyte on kit plate 1, C2 = A2 / B is the correction factor for an analyte on kit plate 2, and so on.
The metabolite concentrations are finally normalized by dividing each concentration value by the correction factor C, e.g. c(analyte, plate1)normalized = c(analyte, plate1) / C1(analyte).
For more information about the normalization algorithms, refer to the Target value normalization and Reference sample normalization sections.