Validation | workflow
The kit run performance is evaluated in the Validation section.
To protect validation status and results from unwanted changes, use Approve plate run to lock results.
Validate plate run
To assess the performance of a kit run, click Validate.
.
Wait until the validation is completed.
Validation features
Within the validation section, the following features are available.
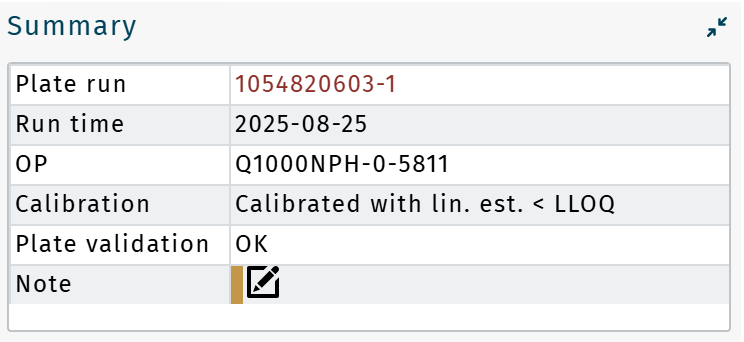
Summary
Plate run: Displays current plate run barcode. Click to load the corresponding data directly in Results.
Run time: Date of MS data uploaded
OP: Plate run OP
Calibration: Plate run calibration status (LC only)
When linear extrapolation < LLOQ was applied, Calibrated with lin. est. < LLOQ is shown.Plate validation: "OK" or "Not OK"
- OK: Plate successfully passed the validation criteria.
- Not OK: Plate did not fully meet the validation criteria. Please review "Analyte results" window.
Note: Click
to open a summary of all validation notes, including a summary of excluded metabolites or wells.
LOD algorithm
The limit of detection (LOD) for each metabolite within a kit run is calculated from the concentrations in the zero samples based on either the standard deviation (StdDev-based) or median (Median-based).
- StdDev-based: LOD is calculated as median concentration of the zero samples concentrations plus three times the standard deviation of the zero samples concentrations. LOD = median background signal + 3 × standard deviation
- Median-based: LOD is calculated as three times the median concentration of the zero samples. LOD = median background signal × 3
Requirement for StdDev-based algorithm:
If StdDev-based or Median-based are defined as default in the Settings > LOD algorithm, this overrules the predefined LOD algorithm of the method (OP).
Default LOD algorithm
Method default: LOD is calculated according to default settings in the method's OP, "StdDev-based" or "Median-based".
For validated plate runs, the LOD algorithm used is shown, e.g. "Median-based".
Example

- Current LOD algorithm of plate run is "Median-based"
- Default LOD algorithm is "StdDev-based"
- Select whether current algorithm ("Median-based") or new ("StdDev-based") algorithm is used for plate run validation.
LOD evaluation
For LOD evaluation, two options are available and used.
- LOD (calc.), the LOD is calculated using zero sample(s) from the corresponding plate run and the LOD algorithm defined in WebIDQ, see LOD algorithm.
- LOD (from OP), LOD values based on kit validation experiments are used, available in kit document "analytical specifications". These LOD values serve as instrument specific, “minimum” LOD values.
When is “LOD (from OP)” used? If a LOD value could not be calculated for a metabolite, e.g. no noise on corresponding MRM integrated, “LOD (from OP)” instead of "LOD (calc.)" is used.
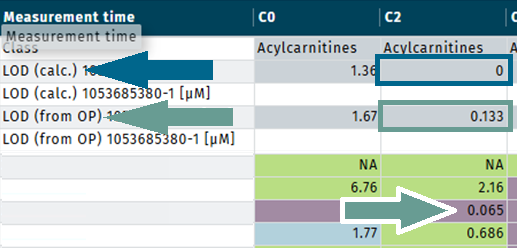
Example for metabolite “C2”
- LOD (calc.) is “0” | blue arrow.
- LOD (from OP) is “0.133” | green arrow.
- For LOD evaluation, “LOD (from OP) = 0.133” is used.
- Concentration “0.065” is flagged as “< LOD” | green arrow.
Information: “LOD (from OP)” and "LOD (calc.)" are available in Results > Data.
LOQ definition
The limit of quantification (LOQ) is defined as
LOQ = LOD × 3.3 .Link sample material with zero samples
If zero samples of different materials are available on one plate run, sample materials need to be linked. Please find an example below.
Zero samples
- PBS
- Isoprop_H2O 80_20
Sample materials
- EDTA plasma
- liver tissue
EDTA plasma is linked with PBS by dragging EDTA plasma to PBS.
liver tissue is linked with Isoprop_H2O 80_20 by dragging liver tissue to Isoprop_H2O 80_20.
This definitions are used for limit of detection (LOD) calculations.
- The zero sample PBS is used for LOD calculations of EDTA plasma samples.
- The zero sample Isoprop_H2O 80_20 is used for LOD calculations of liver tissue samples.
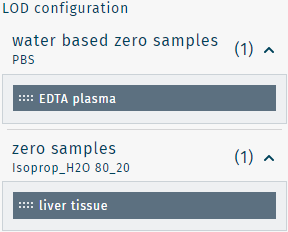
Plate must be validated again after changing zero samples for new the LOD(s) to be applied
Plate view
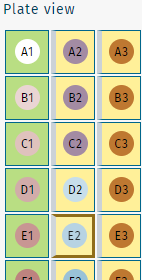
To assess the performance of a plate run, select a QC sample from the Plate view.
The size and level of detail in the plate view is adjustable.
small size
sample IDs shown
sample barcodes shown
large size
- Select a sample, e.g. QC2 on position E2, by clicking on the corresponding well.
- Validation results are displayed in the tables Analyte results and ISTD results.
Coefficient of variations (CVs) are calculated as percentages when at least three QC well replicates are available per plate run.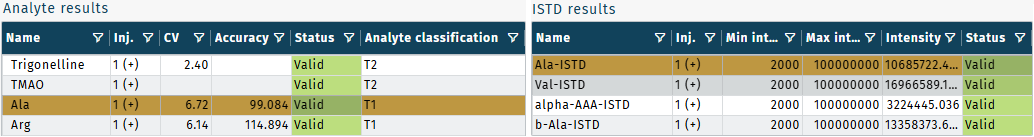
CV = standard deviation / mean concentration * 100Accuracies are calculated for quantitative metabolites only, analyte classification T1.
- Accuracies are calculated for metabolites of analyte classification "T1"
- CVs are calculated for metabolites of analyte classification "T1" or "T2"
- Validation results are illustrated in the Analyte results and ISTD results graphics.
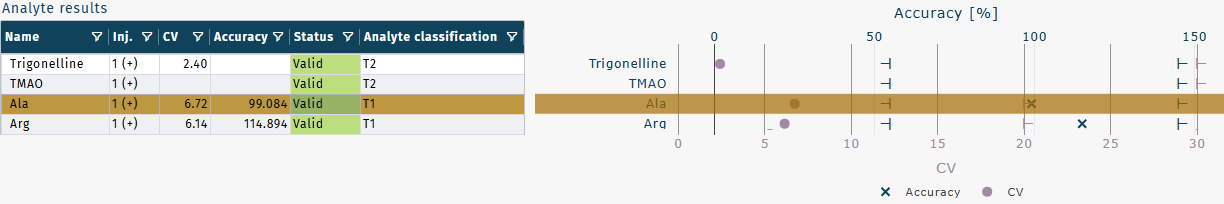
CVs are shown as
.
Accuracies are shown as
.
Acceptance ranges are shown as
in the corresponding color of
or
.
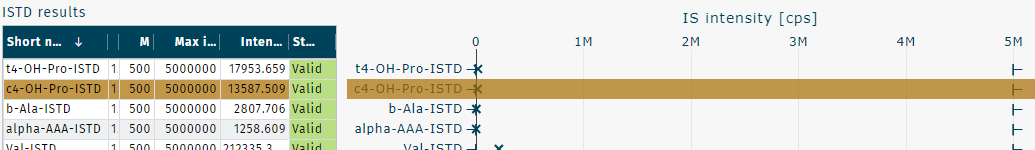
ISTD intensities are shown as
Acceptance ranges are shown as
- Concentrations of samples and counts of internal standard (ISTD) are validated based on acceptance range criteria, resulting in defined validation statuses.
- QC status:
and
are available.
- Calibration standard status:
is available.
- 7-point calibrated metabolites quantification range: LLOQ and ULOQ are the lowest and highest enabled calibration standard level. Calibration standards can be enabled and disabled, see Disable calibration standard levelExample: Cal2 - Cal7 are enabled. LLOQ = concentration of Cal2, ULOQ = concentration of Cal7.
Chromatograms
Chromatograms to a corresponding well can be displayed.
- Click on one well, e.g. QC2 on position E2.
- Select an analyte, e.g. Ala.
Analyte classification
- Results from all biocrates kits are quantitative. The default unit is µmol/L.
- Defined metabolites are quantified using a seven-point calibration based on the calibration standards. The calibration is performed in Quantification > Calibration.
- Other metabolites are quantified using a one-point calibration based on internal standards.
- For details, refer to the kit specific documents "Analytical specifications".
General analyte classification
Analyte classifications are kit specific.
| Analyte classification | Information |
|---|---|
| Type 1 (T1) | Accuracy checked during kit validation. Reproducible (precise) results with low CVs and good accuracy (accurate). |
| Type 2 (T2) | Reproducible (precise) results with low CVs. |
| Qualitative | Acceptance criteria for precission (CV) not met. |
| Not validated (NV) | Validation not possible, e.g. metabolite not available in validation matrix. |
Kit specific analyte classification
Analyte classifications are kit specific, described in the kit specific document Analyte specifications, section "analyte classification".
Validation statuses
- All validation statuses like <LOD, <LLOQ, or >ULOQ refer to the measured concentrations.
- "Measured concentrations" are values, to which no calculations were applied, e.g. data normalization or sample volume different from 10 µL.
- When applying any calculation or normalization to results, concentrations will change but the validation status stays the same.
- Sample material specific LOD: When using zero samples of different materials, e.g. "serum" and "tissue", material specific LOD values are used, according to section Link sample material with zero samples.
- Summary: Validation statuses like <LOD, <LLOQ, or >ULOQ are given before calculation or normalization procedures are performed.
Example
Measured concentration below the LOD received the status
.
Data normalization or other calculations may change the reported concentration in Results, but not the validation status.
Summary: After calculation or normalization procedures were performed, concentrations may be flagged as
but are shown above the LOD.
Values in bold: concentration and status do not match. After normalization, concentration of sample A is shown above the LOD but the status remains
.
Sample A Status Sample B Status Measured concentration 0.122 0.198 Normalized concentration 0.433 0.322 LOD: 0.150
LLOQ: 0.200 Unit: µmol/L
Analyte results
Validation status of concentration.
concentration is within the quantification range, see kit specific document "Analytical specifications"
concentration is below the limit of detection (LOD)
concentration is below the defined "height threshold" (FIA data only)
concentration is below the lower limit of quantification (LLOQ), but above the LOD
concentration is above the upper limit of quantification (ULOQ)
CV is outside the acceptance range
accuracy is outside the acceptance range
intensity of the internal standard (ISTD) is outside the acceptance range
ISTD results
Validation status of internal standard intensity.
intensity is within the acceptance range
intensity of the internal standard (ISTD) is outside the acceptance range
Blank
ISTD signals (MRMs) are checked exclusively.
below upper intensity (cps) acceptance limit
if above intensity (cps) acceptance limit
Approve plate run
Approve plate run requires WebIDQ core+.
- To protect validation statuses and results from unwanted changes, click Approve plate run. WebIDQ functionalities which would result in integration, validation status, or results changes are now disabled.
- To allow changes of a plate run again, click Unapprove.
Enable or disable the functionality approve plate run in the settings.
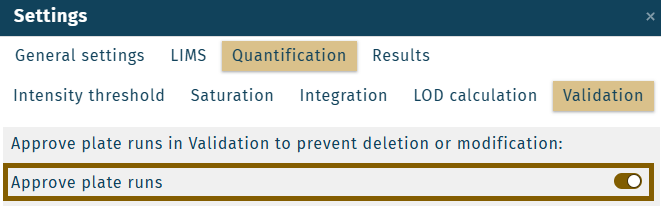
Disabling the setting "Approve plate runs" resets all approved plate runs to unapproved!
Changing the option "Approve plate runs" in the settings requires the WebIDQ user role admin.
To validate a plate run: