Integration | features LC
Integrate
Integration windows are defined in section Integration workflow LC. The WebIDQ quantification algorithm identifies peaks within defined integration windows. After completing the adjustment of all integration windows of a kit plate, click Integrate.
The start and end points of peak integrations can be adjusted globally for all integration procedures, available in Settings > Quantification > Integration.
Discard settings
All injection/well specific integration windows (dashed lines) defined by the user within the selected plate run are replaced with the default integration windows.
- Select a plate run
- Click Discard settings
Discard settings replaces all metabolite or sample specific integration windows with their default values (solid lines), defined for injections/wells of a plate run.
Apply as default
Integration windows from a selected injection/well can be stored for further plate run integrations (LC part of a kit).
- Select an injection/well from the table Wells, e.g. QC2, well 50.
- Click Apply as default.
- Integration windows of the selected well, e.g. QC2, are stored for the next integration of a plate run.
- If no plate run specific integration windows are defined, all default integration windows are replaced with the ones from the selected well.
- Injection/well specific integration windows are not changed (See discard settings above).
To replace injection/well specific integration windows by windows from one well or a CSV file, see Import integration windows, follow this procedure.
- Click Discard settings
- Select one well of the kit plate, e.g. QC2, well 50.
- Click Import integration windows
- While in this example QC2, well 50, is selected, click Apply to plate run
Apply to plate run
Integration windows of a selected injection/well can be applied as the integration windows for the whole plate run (LC part of a kit).
- Select an injection/well from the table Wells, e.g. QC2, well 50.
- Click Apply to plate run.
- Integration windows of the selected well, e.g. QC2, are used for all injections/wells of the currently selected plate run.
- Injection/well specific integration windows are not changed (See discard settings above).
Export integration windows
Integration windows of a selected injection/well can be exported.
- Select an injection/well from the table Wells.
- Click
to export the integration windows of a selected sample, e.g. QC2, well 50.
The export file format is .csv.
Only injection/well specific integration windows can be exported.
Import integration windows
Integration windows can be imported from a WebIDQ compatible .csv file.
- Select an injection/well from the table Wells, e.g. QC2, well 50.
- Click
to apply integration windows to a selected injection/well, e.g. QC2, well 50.
Imported integration windows from a .csv file replace all integration windows of the currently selected injection/well, e.g. QC2, well 50. Integration windows with were defined manually for one sample are not replaced.
Export MS instrument methods - MxP® Quant 1000 and MxQuant kits
Adjusted retention times can be exported directly to instrument methods for MxP® Quant 1000 and MxQuant PITC and NPH methods.
For each method to be exported: In the table “Wells” select a reference sample with updated retention times and in the table “Metabolites” any metabolite, e.g. NPH - Uric acid.
Click Export MS instrument methods.
In the pop-up, define the Window extension in seconds. The window extension will extend the time on either side of the WebIDQ defined integration by the specified amount for the exported MS instrument method.
Recommendation: 8 sec.
- Click Export.
- Acquisition windows for the instrument method will be downloaded, e.g.MS instrument method KitName-0-xxxx MS vendor software (8 sec).xxx
Refer to Technical guide - RT adjustment for updating instrument methods with downloaded acquisition windows.
Integration windows features
Integration window information is shown in tables Wells and Metabolites in a specific column Info.
For integration window status information details, hover with the mouse over an integration window information.
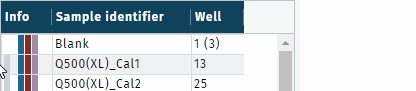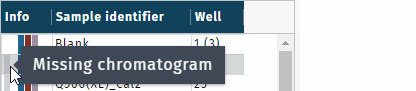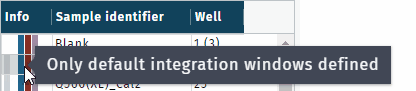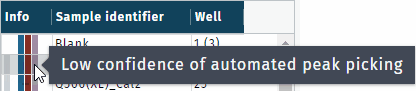
- gray, missing chromatogram(s)
- ochre, missing integration window(s)
- blue, default integration window in use
- red, removed integration windows in use
- purple, RT windows from auto peak picking with low confidence value
- no status, specific integration window in use
Default integration window = same integration window for all samples of a plate run
To find integration window information, define a filter.
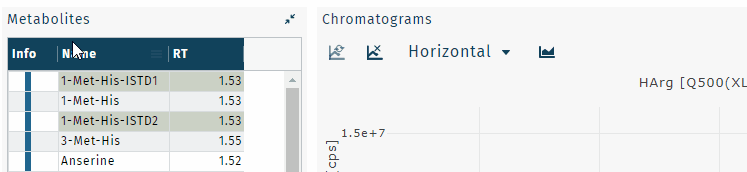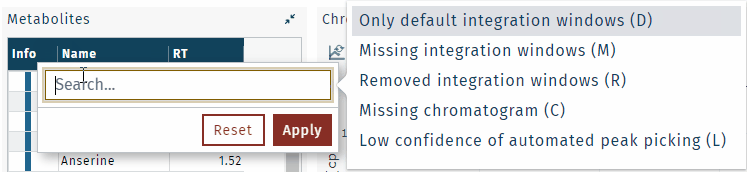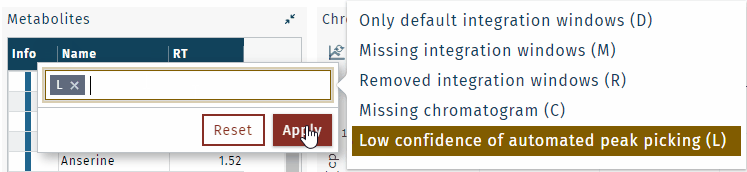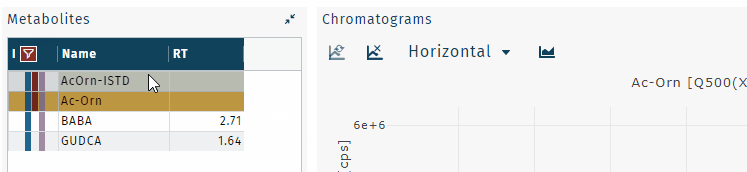
Integration windows of low confidence, highlighted in purple, need to be checked manually.
Chromatogram window features
To adjust the peak integration, several options are available in the Chromatograms panel. Depending if one or multiple injections/wells are selected in the table wells and the position of the zoom slider, adjustments are either applied to one or multiple chromatograms.
| Applied to one chromatogram | Applied to multiple chromatograms |
|---|---|
| Zoom toggle switched to 1, 2, 3, 4, or 5 e.g. | Zoom toggle switched to All |
 |
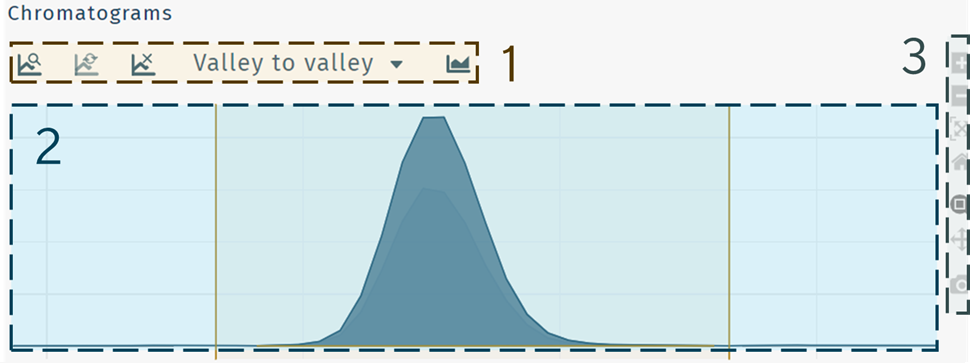
1 peak integration options
Peak integration options
| Find peak | Reset to default integration window | Remove integration window |
|---|---|---|
| Perform "Auto peak picking" | Reset to Default integration window | Apply no peak integration |
Defines peak integration options.
- Valley to valley Peak integrated from left to right curve minimum.
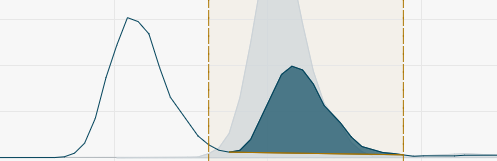
- Horizontal Peak integrated from left to right curve minimum down to baseline.
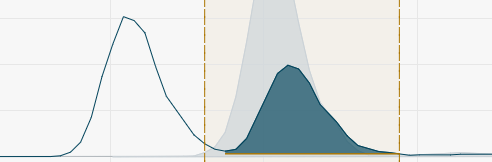
- Manual User defined peak integration width.
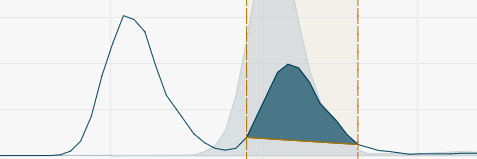
To apply manual changes of peak integration windows, click Reintegrate or press Enter.
Peak display options
Displaying of peaks can be adjusted by one of these options.
| Peak option | Explanation |
|---|---|
| Zoom in / Zoom out | increase or decrease peak size |
| Autoscale | shows all peaks - for metabolites "Reset axes" is recommended |
| Reset axes | best view of selected peak |
| Zoom | zoom in or out using the mouse pointer |
| Pan | move view with mouse pointer |
| Download plot as a png | save view as PNG file |
Examples
Zoom in
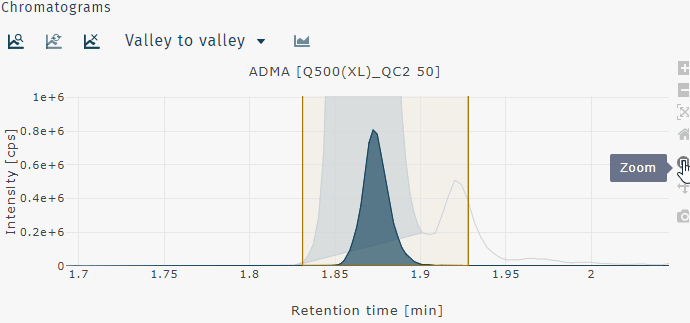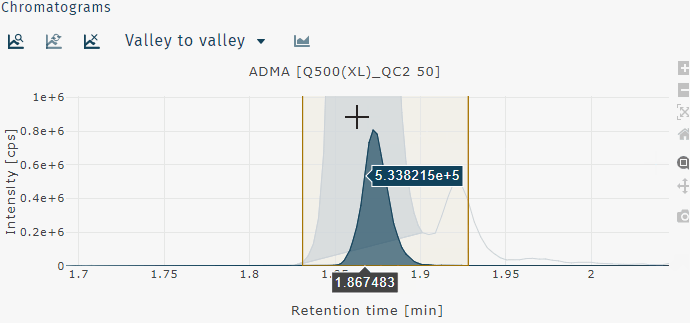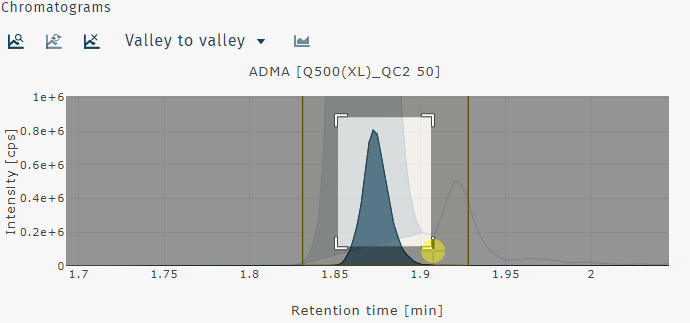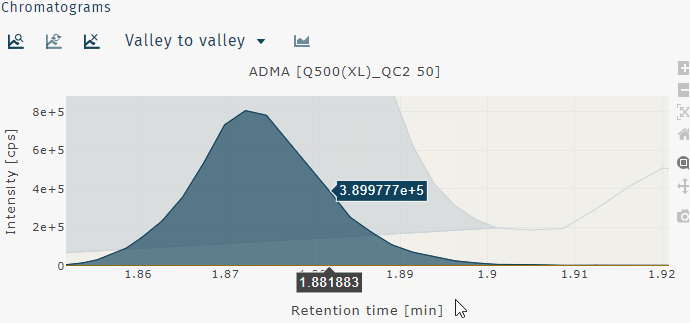
Zoom in can be performed by clicking and draging a box to zoom on that region of the chromatogram or by clicking and dragging across the chromatogram horizontally or vertically to zoom on just the x- or y-axis, respectively.
Zoom out
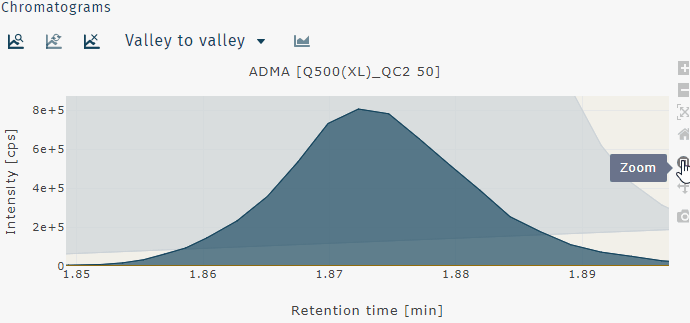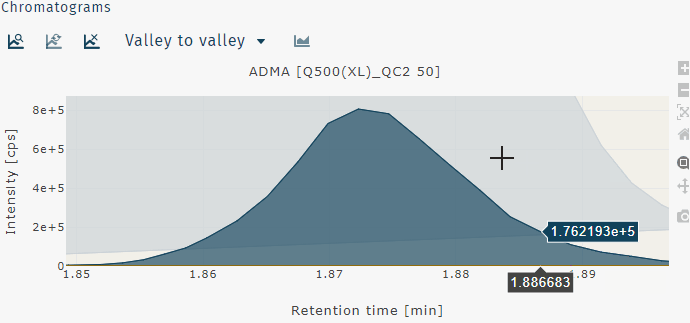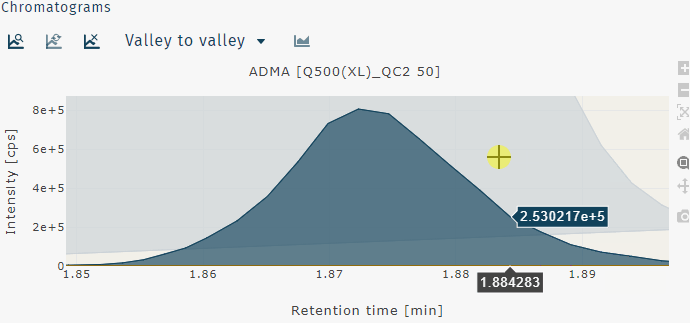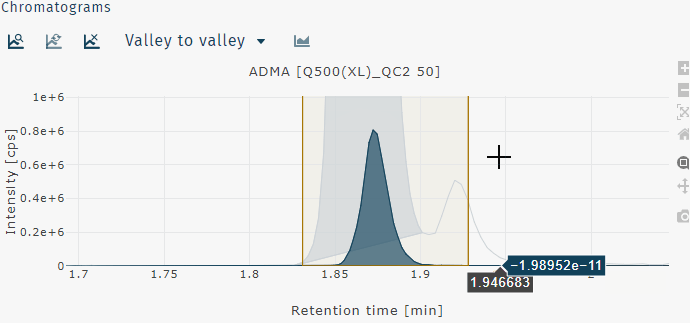
Zoom out can be done by a double left-click.
Reset axes
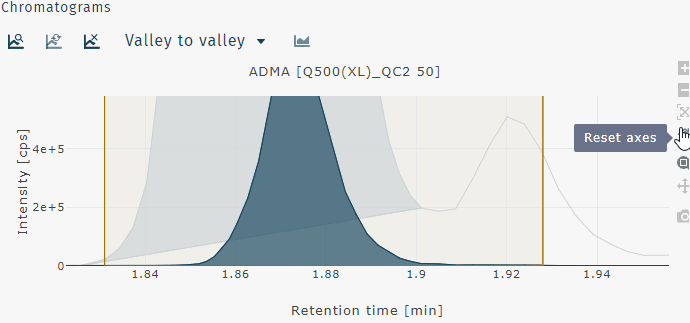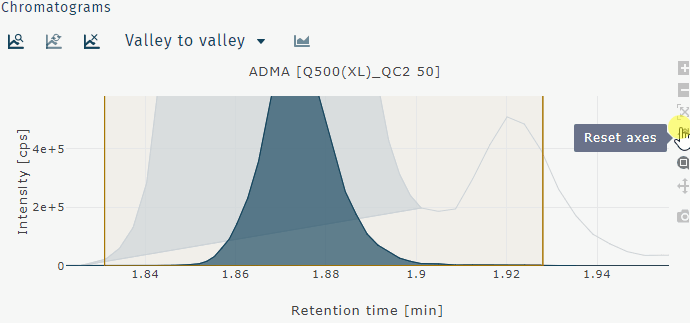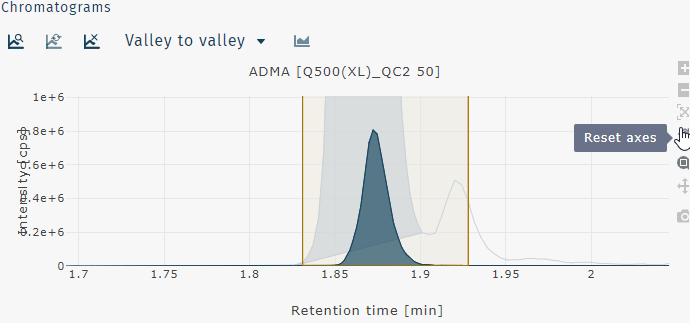
Pan
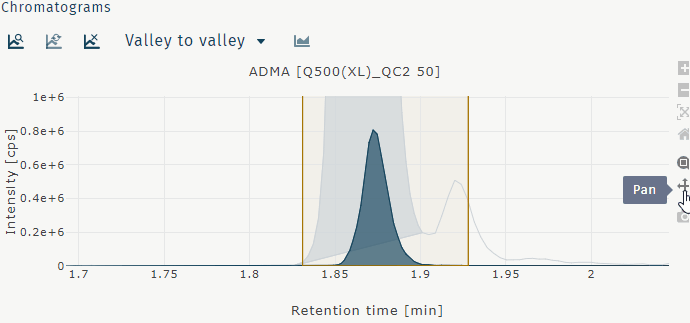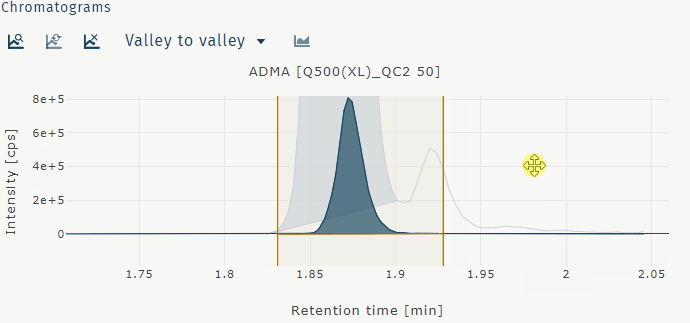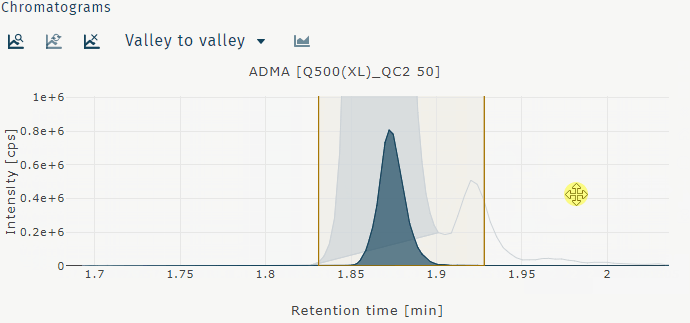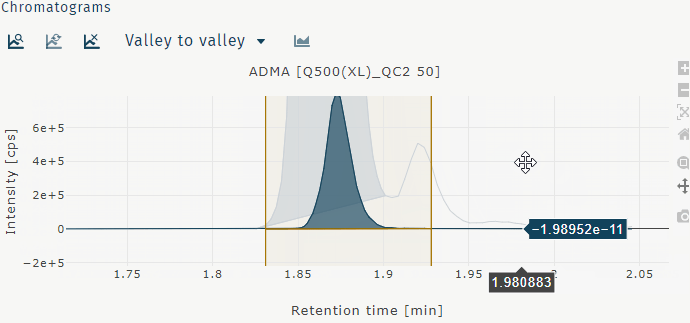
Chromatogram illustration options
Switch between different illustration options by selecting samples from the table wells, selecting a metabolite from the table metabolites, and using the zoom slider.
| Table "wells" | Table "metabolites" | Zoom |
|---|---|---|
Overlay chromatograms | all wells
Overlay all chromatograms of a plate run.
| Table "wells" | Table "metabolites" | Zoom |
|---|---|---|
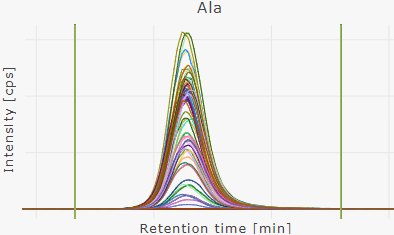
If all wells of a plate run are selected, integration windows are defined as "plate default integration windows".
Default integration window = same integration window for all samples of a plate run.
Overlay chromatograms | multiple wells
Overlay multiple chromatograms of a plate run.
| Table "wells" | Table "metabolites" | Zoom toggle |
|---|---|---|
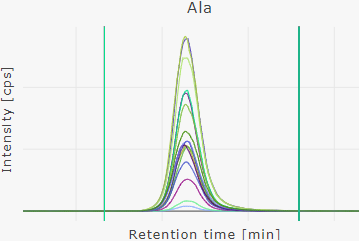
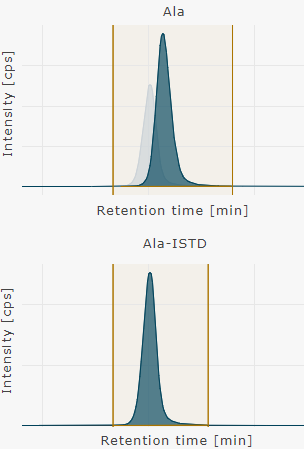
Show individual chromatograms | one well
Show chromatograms for the selected metabolite and its corresponding internal standards (ISTD) of the selected well.
| Table "wells" | Table "metabolites" | Zoom toggle |
|---|---|---|
Show individual chromatograms | two wells
Show chromatograms for the selected metabolite and its corresponding internal standards (ISTD) of the selected wells.
| Table "wells" | Table "metabolites" | Zoom toggle |
|---|---|---|
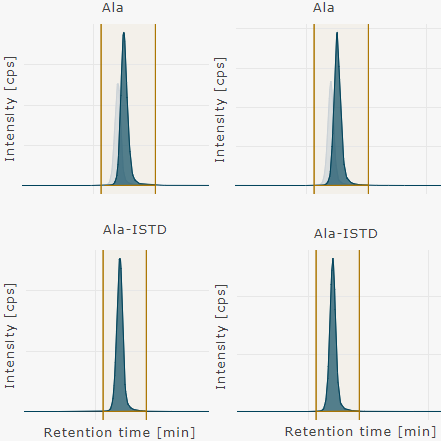
Three, four, and five wells can be shown together accordingly.
Show TIC
Show the TIC for a selected well.
Select a well and any metabolite.
Table "wells" Table "metabolites" Activate Show TIC.
Align RT shifts
If RT shifts occurred during a plate run, to overlay all chromatograms of one metabolite activate Align RT shifts.
| No "align RT shifts" | With "align RT shifts" |
|---|---|
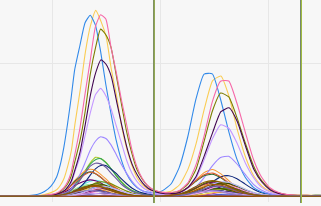 | 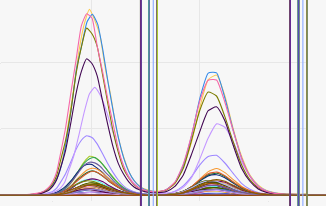 |
The following options are accessible through the menu icon:
 |
|
Align ISTD RT axes
Aligns chromatograms for analyte and internal standard when retentiom times differ. Example GUDCA:
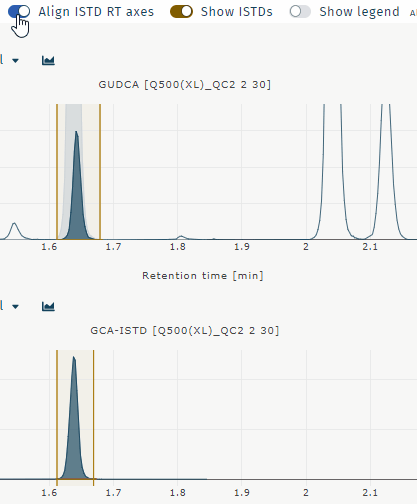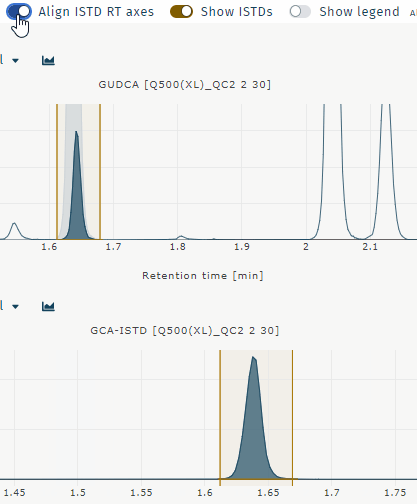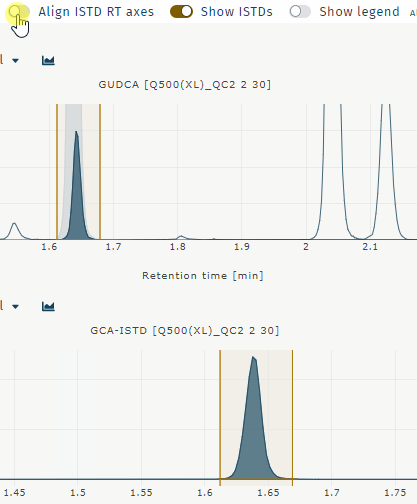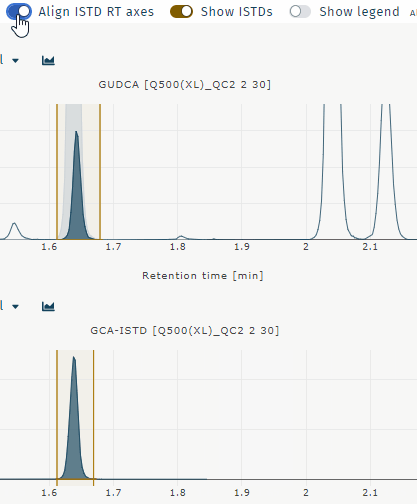
Show ISTDs
Show or hide ISTD chromatogram.
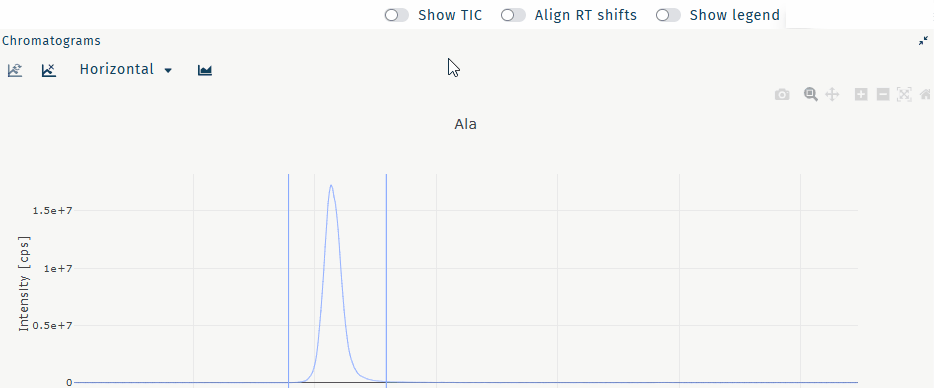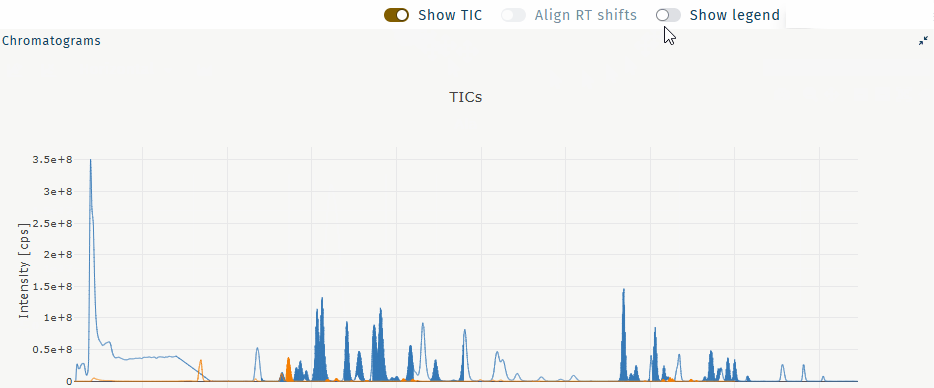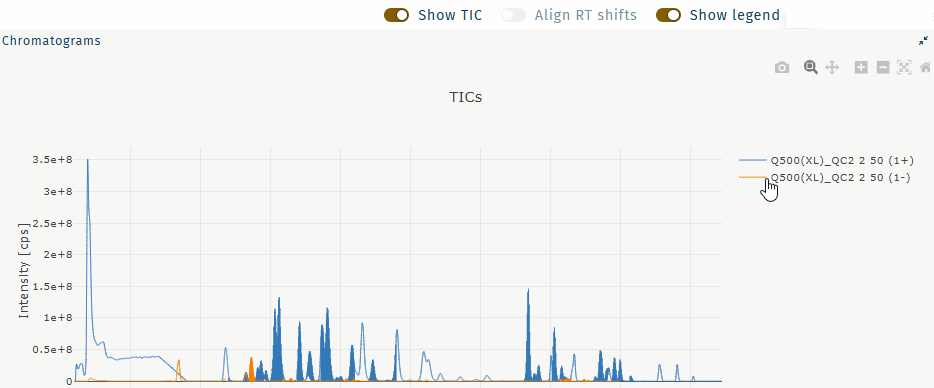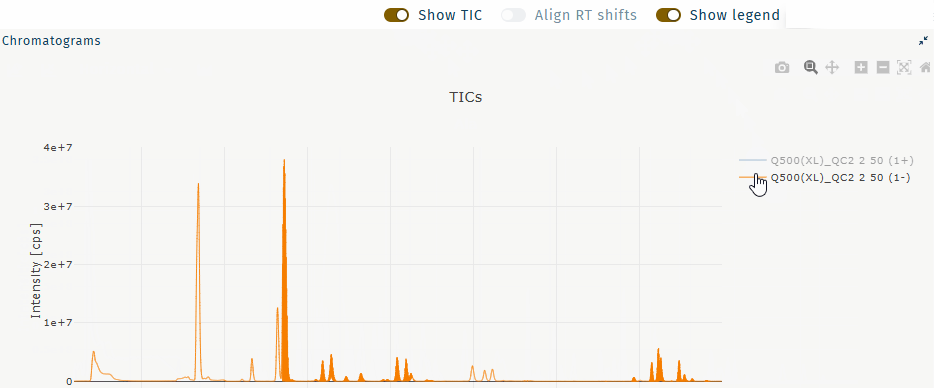
Show legend
Show or hide the legend.
By defining an integration window, quantification is not performed!